MOLECULAR DYNAMICS SIMULATIONS ANALYSIS WITH MOLECULAR PROBES (WEBINAR)
Recorded movie from the first webinar on 23rd SEPTEMBER 2020
ABOUT THE WEBINAR
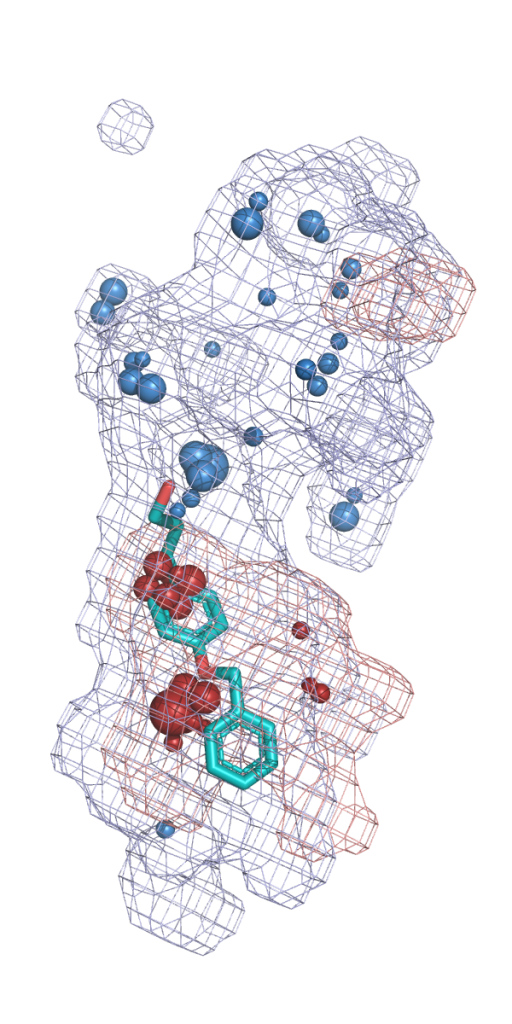
Molecular dynamics simulations are widely used as a versatile research method. Many additional tools were developed to facilitate and 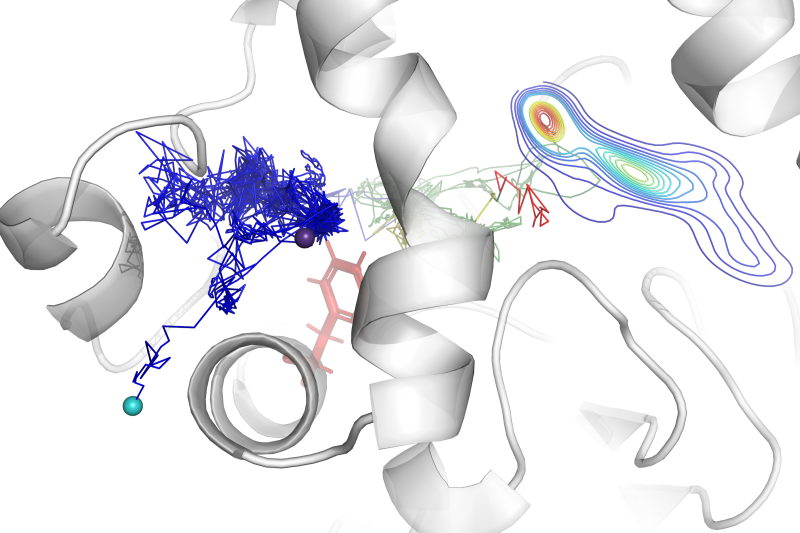 extend analysis of MD simulations. AQUA-DUCT is one of the unique tools, that reverses the standard approach of describing the evolution of macromolecules’ dynamics. Instead of their atoms’ movement analysis, it enables an investigation from the perspective of “intramolecular voids”. To achieve this goal, AQUA-DUCT samples macromolecules’ dynamics employing molecular probes – small entities in simulations (most frequently water molecules, but it could be any solvent, co-solvent, ions, or other ligands). The small molecules are used as specific chemical probes. The mixed “ligands-tracking” and “local-distribution” approaches implemented in AQUA-DUCT allow discrimination between functionally relevant compartments, and provide direct access to information about the active site, potential hot-spots, functional residues (gates, anchoring residues, filters, etc.), the network of internal transportation pathways, and functional voids and cavities. Besides that, it can facilitate the drug design process.
extend analysis of MD simulations. AQUA-DUCT is one of the unique tools, that reverses the standard approach of describing the evolution of macromolecules’ dynamics. Instead of their atoms’ movement analysis, it enables an investigation from the perspective of “intramolecular voids”. To achieve this goal, AQUA-DUCT samples macromolecules’ dynamics employing molecular probes – small entities in simulations (most frequently water molecules, but it could be any solvent, co-solvent, ions, or other ligands). The small molecules are used as specific chemical probes. The mixed “ligands-tracking” and “local-distribution” approaches implemented in AQUA-DUCT allow discrimination between functionally relevant compartments, and provide direct access to information about the active site, potential hot-spots, functional residues (gates, anchoring residues, filters, etc.), the network of internal transportation pathways, and functional voids and cavities. Besides that, it can facilitate the drug design process.
The webinar will provide an opportunity to learn basic and advanced analysis performed with AQUA-DUCT. Participation is free of charge.
Requirements
At least basic knowledge of Linux system and PyMol
Knowledge of classical molecular dynamics simulations (any software)
Literature
AQUA-DUCT 1.0: structural and functional analysis of macromolecules from an intramolecular voids perspective. Magdziarz T., Mitusińska K., Bzówka M., Raczyńska A., Stańczak A., Banas M., Bagrowska W., Góra A., Bioinformatics 2019, 1–3. OPEN ACCESS
AQUA-DUCT a ligands tracking tool. Magdziarz T., Mitusińska K., Gołdowska S., Płuciennik A., Stolarczyk M., Ługowska M., Góra A., Bioinformatics 2017, 33 (13): 2045-2046. OPEN ACCESS
Examples of AQUA-DUCT application
Bzówka M. et.al. Molecular Dynamics Simulations Indicate the COVID-19 Mpro Is Not a Viable Target for Small-Molecule Inhibitors Design OPEN ACCESS
Subramanian K., et al., Distant Non-Obvious Mutations Influence the Activity of a Hyperthermophilic Pyrococcus furiosus Phosphoglucose Isomerase. Biomolecules (2019) 9(6), 212. OPEN ACCESS
Subramanian K., et al., Modulating D-aminoacid oxidase (DAAO) substrate specificity through facilitated solvent access. PLoS ONE (2018) 13(6):e0198990. OPEN ACCESS
Mitusińska K., et al., Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking. Biomolecules (2018) 8(4), 143. OPEN ACCESS
PROGRAMME
AQUA-DUCT – general information, functionality, workflow
Introduction to AQUA-DUCT calculations
Molecular probes applicability in protein engineering
Molecular probes applicability in drug design
REGISTRATION
REGISTRATION IS OVER.
ORGANIZERS
![]()
The Tunneling Group was established in June 2014 at The Biotechnology Centre as an independent research group.
The main activity of the group borders the fields of molecular biology and computational chemistry. We are using an advanced theoretical approach to investigate the properties of various enzymes, to design biologically active compounds, and to help experimentalists interpret their results. We are also developers of the scientific software, AQUA-DUCT and BALCONY. More about group activity on the official site.
Contact
Artur Góra
Email: a.gora@tunnelinggroup.pl
Phone: (+48) 32 237 16 59
or AQUA-DUCT team
Email: info@aquaduct.pl
