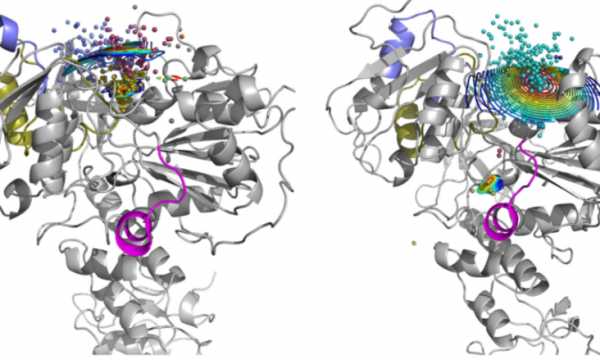
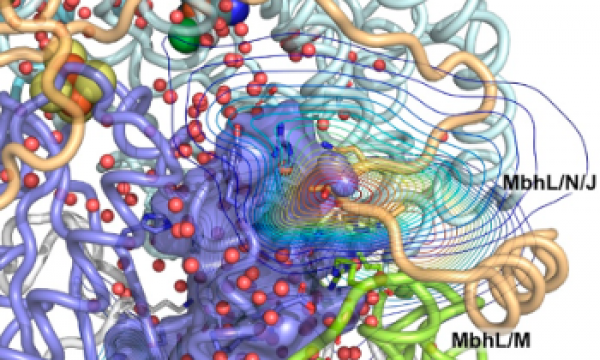
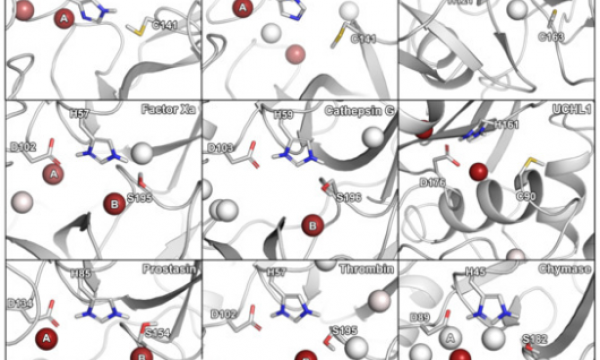
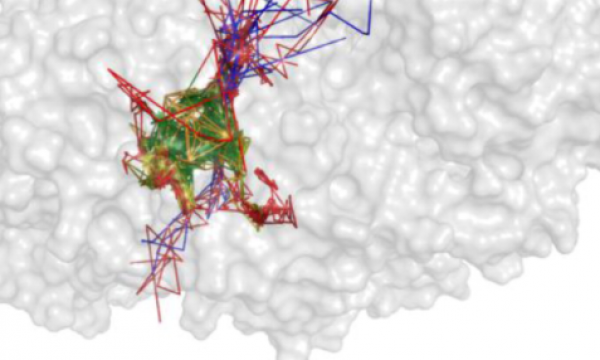
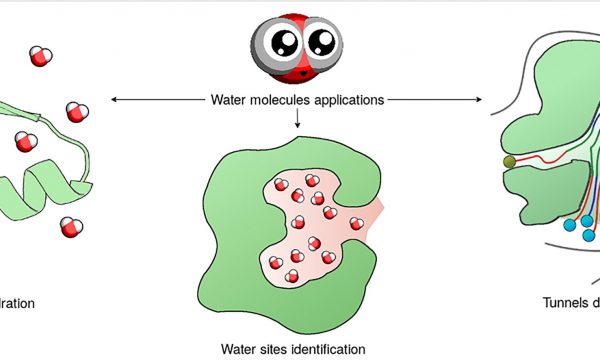
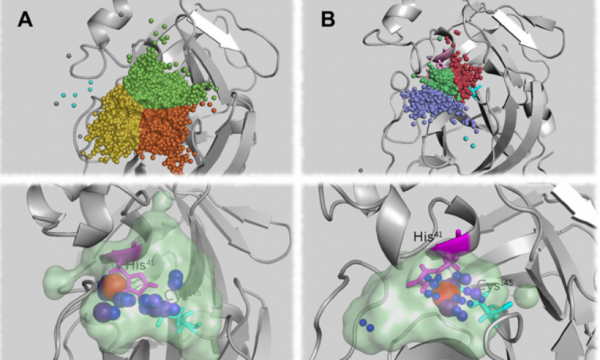
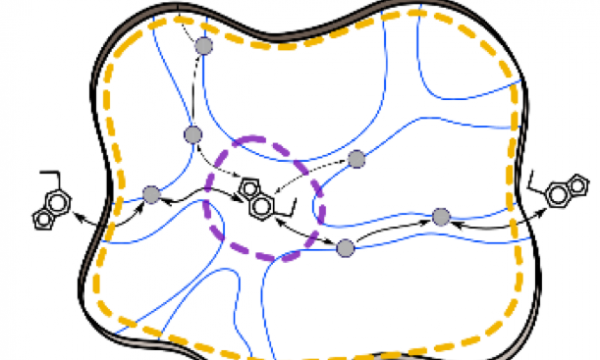
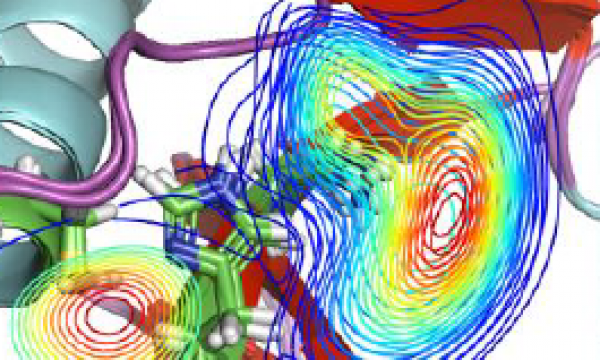
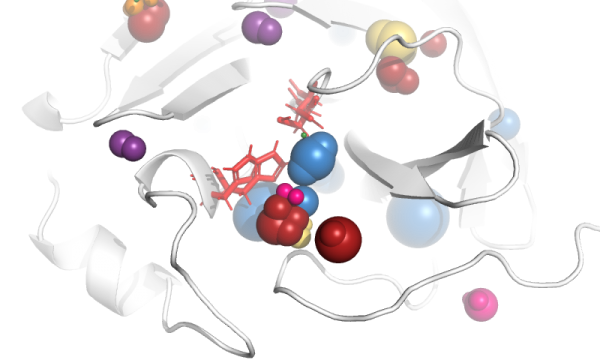
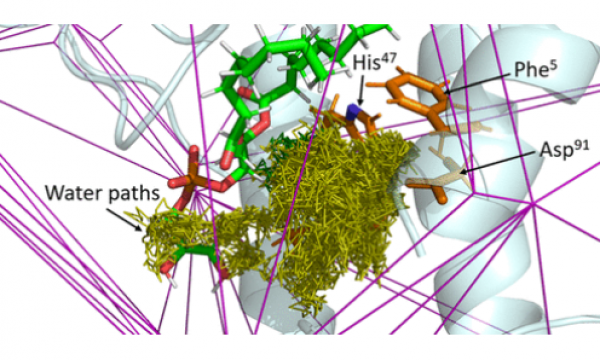
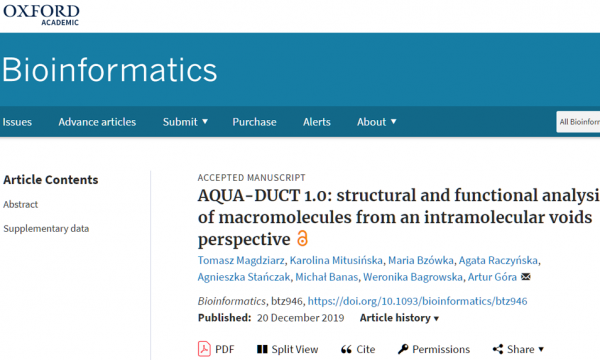
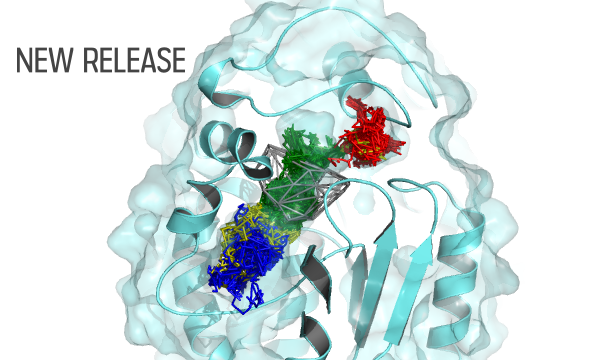
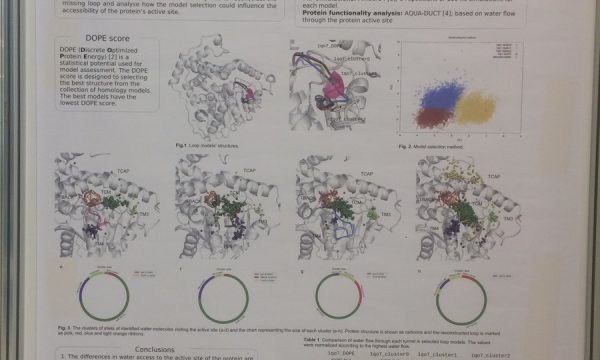
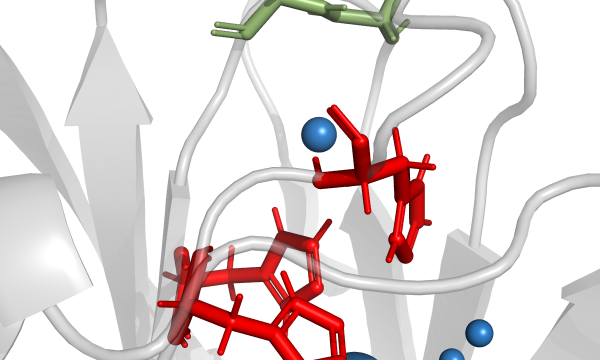
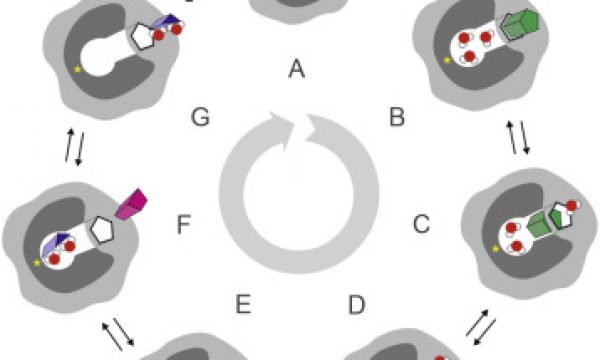
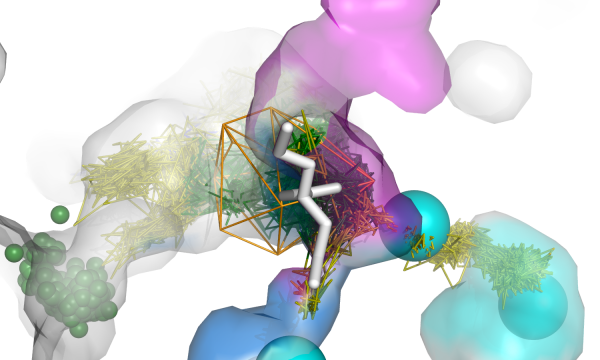
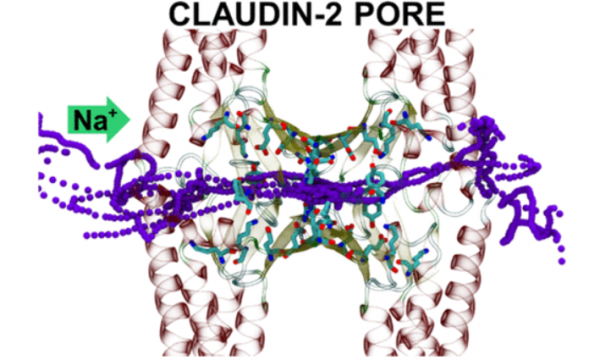
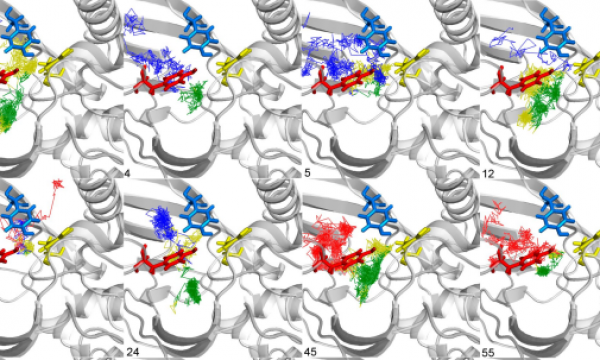
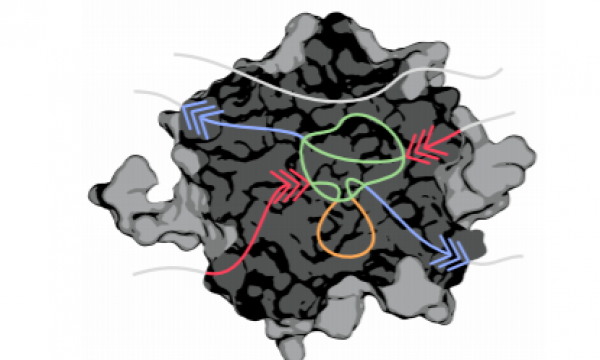
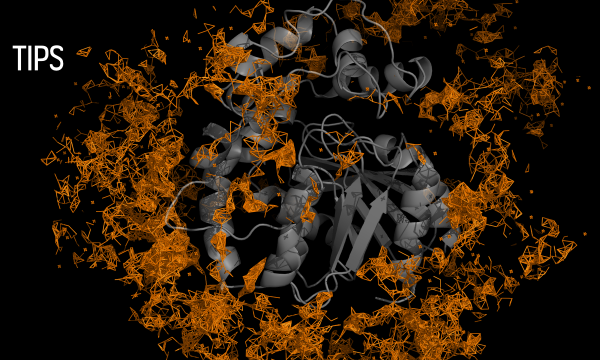
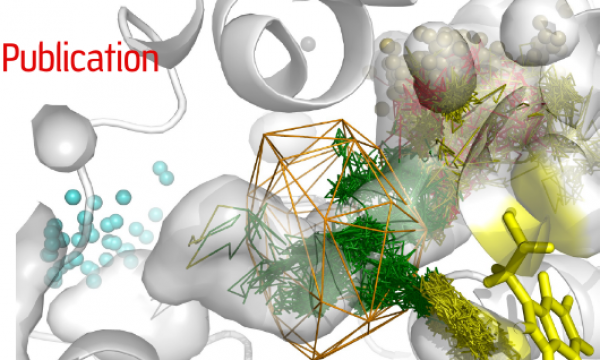
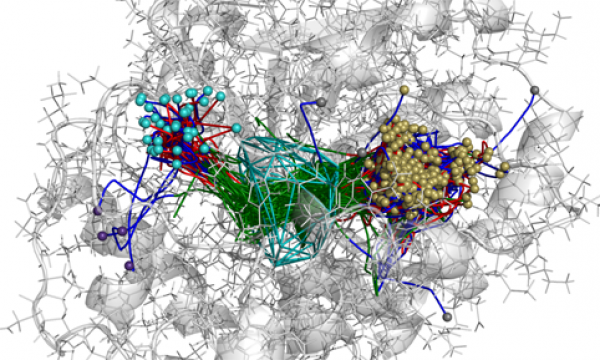
AQUA-DUCT was presented during Course Applied Enzymology organised by Wageningen University
An excellent possibility to get insights how AQUA-DUCT can help with enzymes engineering was presented by Agata one f the AQUA-DUCT developer during poster session and pitch talk at Wageningen.