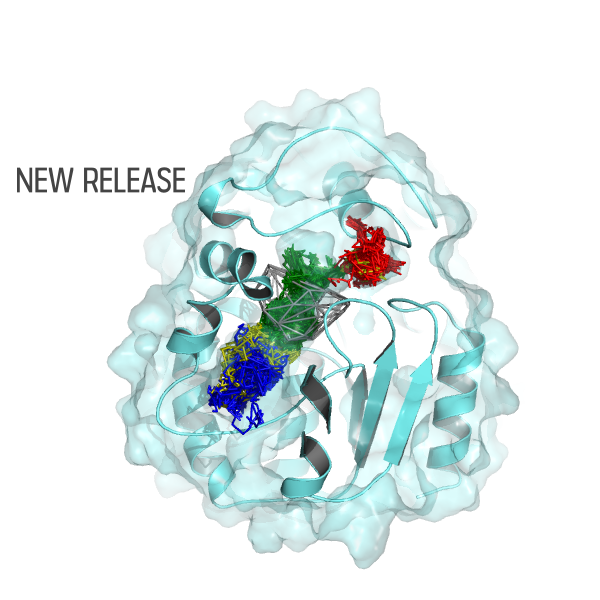
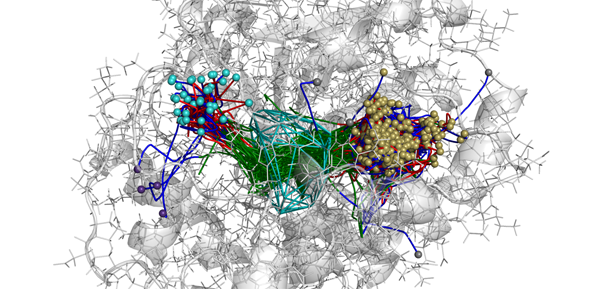
AQUA-DUCT – Release 1.0.0 is available
New version of AQUA-DUCT is available (Release 1.0.0) The new version of AQUA-DUCT is combining information from small ligands tracking and from small ligands local distribution. New modes of AQUA-DUCT MD data processing: normal, sandwich, time-window and consolidator mode. Pond module for hot-spots identification facilitating protein engineering and drug design, Energy barrier approximation. Graphical User…